Vebo đứng đầu trong danh sách các trang web cung cấp dịch vụ xem bóng đá trực tuyến, thu hút lượng lớn người hâm mộ thường xuyên ghé thăm mỗi ngày.
Tại Vebo TV, chúng tôi phát sóng hầu hết các giải đấu bóng đá đỉnh cao trên toàn thế giới, với chất lượng hình ảnh sắc nét không khác gì trên màn hình TV. Ngoài tên gọi Vebo TV, trang còn được biết đến với nhiều tên gọi khác như: Vebo, Về Bờ TV.
Vebo TV mang đến trải nghiệm xem bóng đá trực tiếp hoàn toàn miễn phí và cung cấp thông tin bổ ích về bóng đá. Nếu bạn đang phân vân chọn lựa trang web nào để theo dõi trực tiếp bóng đá, VeboTV là sự lựa chọn không thể bỏ qua.
Giới thiệu về Vebo TV – Trang web trực tiếp bóng đá uy tín hàng đầu hiện nay
Vebo TV là một nền tảng trực tiếp bóng đá mới nổi, ra đời để đáp ứng nhu cầu xem bóng đá trực tuyến đang tăng cao ở Việt Nam.

Với đội ngũ chuyên gia hàng đầu, Vebo TV đã nhanh chóng trở thành điểm đến hàng đầu của người hâm mộ. Mục tiêu của chúng tôi là cung cấp trải nghiệm xem bóng đá trực tiếp tốt nhất cho người dùng trên mọi thiết bị.
Hiện tại, VeboTV liên kết với hầu hết các giải đấu bóng đá hấp dẫn nhất trên thế giới, từ các giải Châu Âu như Cúp C1, Ngoại Hạng Anh, La Liga, giải vô địch quốc gia Đức, giải vô địch các quốc gia Đông Nam Á AFF, U23 Asian Cup, SeaGame.
Bên cạnh đó, chúng tôi cập nhật thông tin đội hình, phong độ và thành tích đối đầu của các đội bóng trước mỗi trận đấu.
Vebo TV hoạt động dưới hình thức phi lợi nhuận, không thu phí người dùng mà chỉ hiển thị quảng cáo để duy trì hoạt động. Ngoài việc cung cấp dịch vụ xem bóng đá trực tiếp, chúng tôi còn đưa ra nhiều thông tin bổ ích khác như tin tức, kết quả, lịch thi đấu và bảng xếp hạng bóng đá.
Tầm nhìn của Vebo trong việc cung cấp dịch vụ xem bóng đá trực tiếp
Trong bối cảnh công nghệ phát triển không ngừng, người hâm mộ bóng đá ngày nay có thể dễ dàng thưởng thức các trận đấu trực tiếp qua máy tính hoặc điện thoại di động.
Hiểu rõ nhu cầu này, đội ngũ kỹ sư công nghệ thông tin của VeboTV đã rất cố gắng và đầu tư thời gian, công sức và tài năng của mình để tạo ra VeboTV – một nền tảng xem bóng đá trực tiếp
Mục tiêu của chúng tôi không chỉ là truyền tải trực tiếp các trận đấu, mà còn là mang đến cho người hâm mộ trải nghiệm chất lượng và dịch vụ tốt nhất có thể.
Từ khi ra mắt, truc tiep bong da Vebo luôn hướng đến phương châm “Không chỉ là truyền tải trực tiếp các trận đấu, mà tạo ra nhiều trải nghiệm bóng đá thú vị, chuyên nghiệp một cách gần gũi nhất”
Website hiện nay được đầu tư để tăng trải nghiệm xem bóng đá, cải thiện hạ tầng và nâng cao chất lượng hình ảnh và âm thanh, với mục đích tăng trải nghiệm tốt nhất cho người hâm mộ.
Với cam kết và trách nhiệm, đội ngũ chăm sóc khách hàng luôn lắng nghe ý kiến feedback từ những khán giả, từ đó trao dồi kỹ năng và hoàn thiện dịch vụ tại Website.
Trải qua nhiều năm hoạt động, Vebo TV – Xem bóng đá trực tuyến đã từng bước khẳng định vị thế của mình trong lòng người hâm mộ. Thương hiệu này là tâm huyết mà đội ngũ mong muốn được nhiều người theo dõi dành tình cảm cho nó.

Hướng phát triển tương lai của VeboTV xem trực tiếp bóng đá
Xem bong da vebo đặt ra bốn mục tiêu chính:
- Trở thành một trong những kênh phát sóng bóng đá trực tuyến hàng đầu, mang lại trải nghiệm tuyệt vời nhất cho người xem.
- Giúp người hâm mộ được trải nghiệm chất lượng hình ảnh, âm thanh sắc nét nhất
- Người hâm mộ sẽ được tham gia hệ sinh thái mạng xã hội của Brand Vebo
- Đem đến những thông tin nhanh và chính xác nhất về bóng đá.
Sứ mệnh của VeboTV là mang đến những trận đấu bóng đá đỉnh cao và miễn phí cho người hâm mộ. Từ khi mở trang web, đội ngũ luôn nỗ lực để đạt được mục tiêu này.
Với cam kết và nhiệt huyết, chúng tôi luôn lắng nghe và tiếp thu ý kiến của những khán giả nhiệt thành, từ đó biết chỗ nào chưa được để cải thiện.
Các trận đấu bóng đá trực tuyến từ VeboTV, bao gồm các giải đấu hàng đầu như giải Ngoại hạng Anh, vô địch Tây Ban Nha La Liga, vô địch Đức, Ý, Pháp được cập nhật một cách nhanh nhất và cụ thể.
Mỗi trận đấu đều có sự tham gia của các bình luận viên chuyên nghiệp, tạo nên không khí vui vẻ và thú vị cho người xem.
Với Vebo live truc tiep bong da, người dùng không chỉ cần trả phí internet hàng tháng mà còn được tận hưởng trải nghiệm xem bóng đá trực tiếp hoàn toàn miễn phí.
Trang web luôn hướng đến người hâm mộ để cải thiện chất lượng âm thanh hình ảnh và một giao diện thân thiện với người dung trên tất cả các thiết bị (PC, Tablet, Smartphone)
Với phương châm “Customer is God”, Vebo TV – Xem trực tiếp bóng đá ngày càng khẳng định vị thế của mình và trở thành nơi để người hâm mộ có thể giải trí mà vẫn nhận được thông tin uy tín nhất.
Các chuyên mục chính tại VeboTV – Kênh trực tiếp bóng đá
Với sứ mệnh phục vụ cộng đồng người hâm mộ bóng đá và các mục tiêu đã đề ra như trực tiếp bóng đá hôm nay, Website không ngừng phát triển và hoàn thiện. Ngoài các trận đấu bóng đá hấp dẫn, Vebo TV còn cung cấp các phân loại sau đây:
Bảng xếp hạng bóng đá trực tiếp các giải đấu nổi tiếng
Đây là nơi mà người xem có thể tìm hiểu thông tin về bảng xếp hạng bóng đá trực tiếp hôm nay tại Vebo TV. Thông qua đó, người hâm mộ có thể đánh giá sức mạnh của hai đội một cách tương đối.
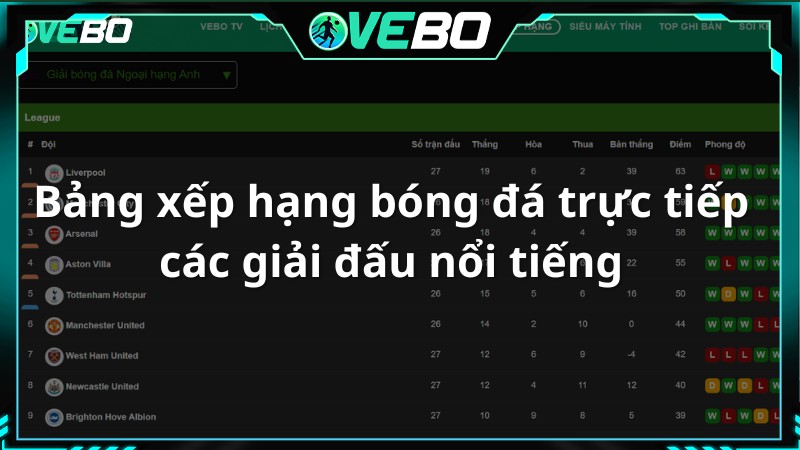
Vebo live – Kênh xem bóng đá trực tuyến cập nhật thông tin bảng xếp hạng nhanh chóng, điều chỉnh vị trí, hiệu suất, điểm số và thành tích của mỗi đội để phản ánh chính xác tình hình. Thông tin về số trận thắng, hòa, thua, tổng số bàn thắng và bàn thua cũng được cung cấp đầy đủ.
Ngoài ra, phần Bảng Xếp Hạng còn liên kết với nhiều giải đấu mà một đội bóng tham gia, khán giả chỉ cần chọn mục giải đấu và lựa chọn cho mình mục mà mình quan tâm nhất.
Kết Quả Thi Đấu Bóng Đá
Phân mục này cập nhật nhanh chóng kết quả của các trận đấu, giúp người xem dễ dàng theo dõi. Khán giả hoàn toàn có thể tra cứu tỷ số, kết quả trận đấu đã diễn ra của tất cả các giải đấu lớn trên thế giới.
Ngoài tỷ số, trực tiếp bóng đá Vebo TV còn liên kết với Bảng Xếp Hạng của giải đấu tương ứng, giúp người hâm mộ có thể tìm kiếm các thông tin một cách nhanh chóng và tiện lợi.
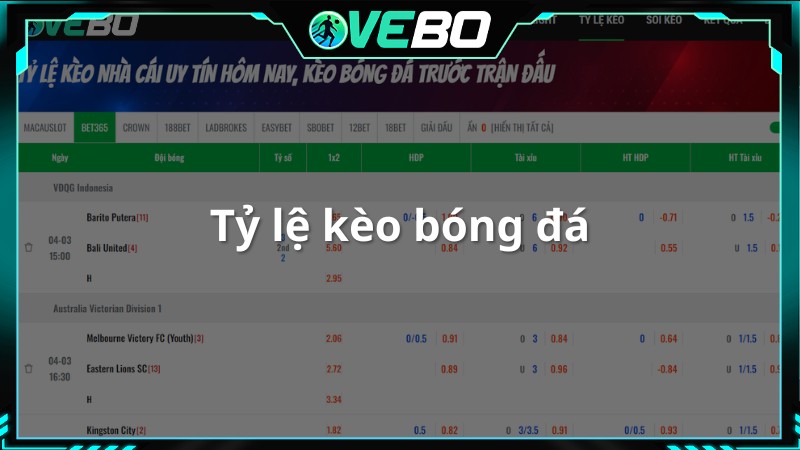
Tỷ Lệ Bóng Đá
Phân mục này cung cấp tỷ lệ bóng đá cùng với thông tin kết quả. Bên cạnh tỷ số, người xem còn có thể xem các tỷ lệ tài xỉu, 1×2, Handicap nếu quan tâm.
Hơn nữa, VeboTV cung cấp địa chỉ cược để người dùng dễ dàng lựa chọn. Các tổ chức được cung cấp bởi website đều là những đơn vị uy tín, đã đảm bảo được chất lượng trước đó về thông tin cũng nhu nhận định bóng đá chuyên sâu trước đó.
Tổng Kết Các Pha Bóng Đỉnh Cao
Sau khi trận đấu kết thúc, bạn có thể dễ dàng theo dõi lại những pha bóng đỉnh cao, truc tiep bong da hom nay, thông qua phần tổng kết.
Tính năng này giúp ích rất nhiều cho việc khán giả theo dõi Vebo không có nhiều thời gian vẫn có thể theo dõi được những highlight (tình tiết hấp dẫn) trong trận đấu.
Điểm mạnh của của trang Vebo xem bóng đá trực tuyến
Chúng tôi tự tin khẳng định rằng Ve Bo TV là lựa chọn hàng đầu cho những ai muốn xem bóng đá trực tuyến một cách mượt mà và không bị giật, lag. Chúng tôi luôn đặt trải nghiệm của người dùng lên hàng đầu, đảm bảo bạn sẽ có những khoảnh khắc thú vị nhất khi xem bong da.
Ngoài ra, kênh Vebo bóng đá còn có những ưu điểm vượt trội so với các trang web khác, bao gồm:
Đa dạng giải đấu
Trực tiếp bóng đá vebotv liên kết để phát sóng một loạt các giải đấu bóng đá hấp dẫn nhất trên toàn thế giới, đảm bảo mang đến sự đa dạng và phong phú cho người xem. Các giải đấu nổi tiếng mà Vebo TV Live phát sóng bao gồm:
- Ngoại Hạng Anh: Giải đấu bóng đá hàng đầu được yêu thích nhất tại Việt Nam.
- Cúp C1: Sân chơi của các đội bóng hàng đầu Châu Âu.
- World Cup: Giải đấu bóng đá lớn nhất và uy tín nhất trên toàn cầu.
- Euro: Giải đấu bóng đá cấp đội tuyển hàng đầu Châu Âu.
- Copa America: Giải đấu danh giá nhất dành cho các đội tuyển Nam Mỹ.
- Serie A: Giải vô địch quốc gia Italy, giải đấu cấp độ cao nhất tại quốc gia này
- Bundesliga: Sân chơi của các đội bóng hàng đầu Đức.
- La Liga: Giải đấu bóng đá uy tín nhất tại Tây Ban Nha.
- Ligue 1: Giải đấu số một quốc gia Pháp, biểu tượng của Les Bleus
- V-League: Giải đấu bóng đá hàng đầu trong nước Việt Nam.
- Các giải đấu cùng cấp độ mà Đội tuyển quốc gia Việt Nam tham gia như: SeaGames, AFF, Vòng loại World Cup, U23 Asian Cup.
Ngoài ra, chúng tôi cũng trực tiếp các giải đấu từ Hàn Quốc, Nhật Bản, Hà Lan và nhiều quốc gia khác, mang đến sự đa dạng và đầy đủ cho người hâm mộ bóng đá.
Nhiều đường link xem bóng đá trực tiếp đáp ứng nhu cầu người xem đông
Khi truy cập Vebo link, mỗi trận đấu được cập nhật với ít nhất 3 đường link xem bóng đá khác nhau, mỗi link có chất lượng hình ảnh khác nhau như HD, Full HD, 4K, để bạn có thể lựa chọn phù hợp với thiết bị của mình.
Chúng tôi cập nhật đường link xem trực tiếp bóng đá trước trận đấu 1 tiếng, đảm bảo bạn có thể truy cập nhanh chóng và an toàn vào xem bóng đá trực tiếp hôm nay. Đường link bong da của chúng tôi luôn đảm bảo không chứa virus hoặc mã độc, mang lại sự yên tâm cho người xem.

Giao diện website thân thiện với người dùng
Chúng tôi hiểu rằng không phải ai cũng thông thạo công nghệ khi xem bóng đá trực tiếp. Vì vậy, giao diện của Ve Bo TV được thiết kế đơn giản và dễ sử dụng, phù hợp với mọi đối tượng người dùng.
Màu sắc nhẹ nhàng và hợp mắt giúp bạn có thể xem lâu mà không bị mỏi mắt. Với giao diện khoa học, việc tìm kiếm và xem trận đấu bạn yêu thích trở nên đơn giản hơn bao giờ hết. Đồng thời, bạn cũng có thể tìm thấy thông tin bóng đá một cách nhanh chóng và tiện lợi.
Chất lượng trận đấu tối ưu
Vebo Tivi cam kết mang đến trải nghiệm xem bóng đá trực tuyến tương đương với việc thưởng thức trên màn hình TV với độ phân giải cao. Là một trong những trang web tiên phong tại Việt Nam áp dụng công nghệ 4.0 trong trực tiếp bóng đá, chúng tôi đảm bảo chất lượng trận đấu như sau:
- Hình ảnh trực tiếp mỗi trận đấu luôn sắc nét, cung cấp lựa chọn đa dạng về độ phân giải như HD, Full HD, 4K,…
- Âm thanh trận đấu sống động và chân thực, không bao giờ bị tắt tiếng trong quá trình diễn ra trận đấu.
- Màn hình xem trực tiếp bóng đá hôm nay được tối ưu hóa với kích thước phù hợp, tương thích với nhiều thiết bị từ máy tính đến điện thoại di động.
Tốc độ mượt mà và ổn định
Chúng tôi đã đầu tư lớn vào công nghệ đường truyền hiện đại nhất để đảm bảo tốc độ mượt mà và ổn định nhất khi xem bong da truc tiep tại Vebo TV. Cụ thể:
- Tải trận đấu chỉ mất vài giây.
- Trải nghiệm xem trận đấu mượt mà và không giật lag từ khi trận đấu bắt đầu đến khi kết thúc.
- Không có hiện tượng giật, đứng hình, lag hoặc mất kết nối trong suốt quá trình xem trận đấu.
Bình luận đầy hấp dẫn
Vebo TV tận dụng sức hấp dẫn của bình luận để làm cho trải nghiệm xem tường thuật trực tiếp bóng đá hôm nay thêm phần thú vị. Chúng tôi đã chiêu mộ các bình luận viên hàng đầu tại Việt Nam, chuyên môn và hài hước, để tạo ra không khí sôi động hơn cho mỗi trận đấu.

Bên cạnh đó, chúng tôi còn mở cửa cho người xem tham gia bình luận, giao lưu và đặt câu hỏi trực tiếp cho các bình luận viên, tạo ra một không gian trò chuyện sôi động và hấp dẫn.
Không quảng cáo làm phiền
Vebo trực tiếp bóng đá cam kết không làm phiền người xem với quảng cáo quá nhiều trong quá trình xem bóng đá trực tuyến.
Chúng tôi luôn tập trung vào trải nghiệm người dùng, chỉ hiển thị các banner quảng cáo nhằm hỗ trợ hoạt động và duy trì website một cách nhất quán. Người xem có thể dễ dàng tắt quảng cáo nếu không muốn xem.
Cập nhật thông tin chi tiết trước trận đấu
Bên cạnh việc cung cấp trực tiếp bóng đá chất lượng cao tại Về Bờ TV, chúng tôi còn cập nhật toàn bộ dữ liệu và thông tin quan trọng liên quan đến trận đấu trước khi nó diễn ra. Mỗi trận đấu được phát sóng trên Về Bờ TV đều được chúng tôi nghiên cứu và cập nhật thông tin như:
- Đội hình dự kiến ra sân của hai đội bóng
- Phong độ gần đây của hai đội.
- Lịch sử đối đầu giữa hai đội.
- Tiềm lực, chất lượng cầu thủ của các đội bóng hiện tại như nào
- Thông tin về sân vận động, trọng tài, thời tiết, vv.
Với những thông tin này, bạn sẽ có cái nhìn chi tiết về cả hai đội trước khi trận đấu bắt đầu.
Lý do bạn nên xem trực tiếp bóng đá tại VeBo TV?
Trên đây là những ưu điểm nổi bật của chúng tôi so với các trang web khác phát sóng bóng đá. Về Bờ TV đang dẫn đầu trong việc phát sóng trực tiếp bóng đá tại Việt Nam.

Do đó, hãy truy cập vào đây nếu bạn muốn xem bất kỳ trận đấu nào với chất lượng tốt nhất. Khi xem bóng đá trực tuyến tại Vebo, bạn hoàn toàn sẽ được:
- Thưởng thức mọi trận đấu từ các giải đấu lớn đến giải đấu cỏ, bao gồm cả các kênh lớn như K+, VTV miễn phí.
- Tận hưởng trận đấu với chất lượng hình ảnh, âm thanh, tốc độ tải trận đấu và kích thước màn hình tốt nhất.
- Không bị làm phiền bởi quảng cáo hoặc popup khi trận đấu bắt đầu, cùng với sự xuất hiện của các BLV hàng đầu.
- Xem trận đấu mượt mà và ổn định nhất nhờ vào đường truyền hiện đại của chúng tôi.
- Sử dụng giao diện được thiết kế khoa học và cập nhật thông tin trước trận đấu như đội hình, phong độ, lịch sử đối đầu một cách đầy đủ.
Những lợi ích trên chính là lí do bạn không nên bỏ qua Vebo khi muốn xem bong da truc tuyen.
Hướng dẫn cách xem bóng đá trực tiếp tại Về Bờ TV
Như đã trình bày, trang web Vebo TV được thiết kế một cách khoa học để việc tìm và xem trận đấu bóng đá trở nên đơn giản hơn bao giờ hết. Đầu tiên, bạn cần chuẩn bị một thiết bị có kết nối mạng như điện thoại hoặc máy tính, có thể là wifi, 3G, 4G, vv. Tiếp theo, khán giả hãy thực hiện các bước sau:
- Truy cập vào tên miền https://shopwestcomb.com/ rồi vào trang chủ của VeboTV
- Chọn mục “Trực Tiếp” từ thanh Menu.
- Tìm giải đấu bạn quan tâm, ví dụ như Champion League.
- Chọn trận đấu muốn xem và chọn đường link phù hợp, sau đó nhấn “Xem Ngay”.
Với những bước đơn giản trên, bạn đã sẵn sàng xem bóng đá trực tiếp tại ve bo tv một cách dễ dàng.
Các tính năng khác trên Vebo
Về Bờ TV không chỉ được đánh giá là trang web phát sóng bóng đá trực tuyến hàng đầu hiện nay mà còn cung cấp một loạt các thông tin bóng đá khác cho người hâm mộ. Đó bao gồm:
- Cập nhật tin tức, sự kiện bóng đá mới nhất trên toàn thế giới hàng ngày.
- Kết quả trận đấu mới nhất cùng thông tin chi tiết.
- Lịch thi đấu các đội bóng lớn hoặc các đội bóng mà bạn quan tâm cùng với bảng xếp hạng cập nhật liên tục.
- Tỷ lệ kèo nhà cái cho tất cả các trận đấu diễn ra trong ngày.
- Nhận định trước trận đấu và tin soi kèo từ các chuyên gia.
- Video highlight giúp bạn xem lại những tình huống đáng chú ý.
- Cập nhật kết quả bóng đá và dữ liệu đội bóng mới nhất.
Với những ai đam mê bóng đá, Về Bờ TV là một nguồn thông tin không thể bỏ qua, nơi bạn không chỉ xem truc tiep bong da mà còn có được các thông tin bổ ích hàng ngày.
Danh sách các trang web xem bóng đá trực tuyến tốt nhất hiện nay
Ngoài Vebo, một trong những kênh bong da truc tuyen được yêu thích nhất hiện nay, dưới đây là một số website khác cung cấp dịch vụ trực tiếp bóng đá với chất lượng tương đối ổn định:
- Xôi lạc TV (XoilacTV2.net | XoilacTV | XoilacTV net | XoilacTV1 | XoilacTV2 | Xoi-lac.TV | Xôi lạc TV)
- 90 Phút TV (90phut.net | 90Phut TV | 90m TV | 90P TV | 91Phut | 91Phut Link)
- Cà Khịa TV (Cakhia.com | Cakhia TV | Ca Khia TV | Ca Khia Link | Cakhia Live)
- Bàn Thắng TV (Banthang.tv | Banthang TV | Banthang.live)
- Mì Tôm TV (Mitom1.tv | Mitom.TV | Mi Tom TV | Mitom TV | Mitom net)
- Vào Rồi TV (Vaoroi.TV | VaoroiTV | Vao Roi TV | Vaoroi TV)
- Thức Khuya TV (Thuckhuya.TV | Thuc Khuya Link | Thuc Khuya TV)
- Ngoác TV (Ngoac.TV | Ngoác TV | Ngoạc TV | NgoacTV)
- VTV (VTV6 | VTV3 HD | VTV9)
- VTC (VTC1 | VTC7)
- K+ TV (K+1 | K+ Phong Cách)
- Thẻ Vàng TV (Thevang.TV | Thevang TV | The Vang TV)
- Sương TV (SuongTV | Sướng TV | SuongTV)
- Thẻ Đỏ TV (Thedo.tv | Thedo TV)
- Bánh Khúc TV (Banhkhuc.TV | Banh Khuc TV)
- Ra Khơi TV (Rakhoi.TV | Rakhoi.link | Ra Khoi TV)
- Sao Kê TV (Saoke.TV | Saoke.link | Ra Khoi TV)
- CheckVAR
Những trang web này cung cấp sự lựa chọn đa dạng cho người hâm mộ bóng đá để theo dõi các trận đấu yêu thích của họ một cách thuận tiện và dễ dàng.
Những lưu ý khi thưởng thức bóng đá trực tiếp trên Vebo TV
Khi truy cập vào Về Bờ TV để xem bóng đá trực tiếp, bạn cần lưu ý các điều sau:

- Đảm bảo kết nối mạng ổn định để trải nghiệm trận đấu mà không gặp phải giật lag.
- Trong trường hợp trận đấu gặp sự cố giật lag, bạn nên tạm dừng và chờ khoảng 3 phút trước khi tiếp tục, thường sẽ khắc phục được tình trạng này.
- Nếu đường truyền gặp sự cố hoặc đường link bị lỗi, bạn có thể chuyển sang link khác để tiếp tục thưởng thức trận đấu.
- Tại Vebo TV, bạn có thể chọn xem ở cả chế độ màn hình dọc và ngang. Tuy nhiên, trên điện thoại, việc xem ở chế độ ngang thường mang lại trải nghiệm tốt hơn.
Với hệ thống link trực tiếp bóng đá miễn phí mà Veboz.net cung cấp, chúng tôi tin rằng bạn sẽ có những giây phút thư giãn và hài lòng nhất khi theo dõi đam mê bóng đá của mình.
Đánh giá kênh Về Bờ TV từ quan điểm của người dùng
Sau thời gian hoạt động, Vebo TV trực tiếp bóng đá đã khẳng định vị thế của mình trong cộng đồng người xem. Sự đánh giá tích cực về chất lượng dịch vụ và các trận đấu mà Về Bờ TV cung cấp liên tục tỏa sáng. Dưới đây là một số phản hồi tích cực từ cộng đồng người hâm mộ dành cho VeboTV.
Việc xem bóng đá trên Về Bờ TV là một trải nghiệm không gây lo ngại. Khác với nhiều kênh phát sóng trực tiếp bóng đá khác, Về Bờ TV không lợi dụng thông tin cá nhân của người dùng với mục đích tài chính.

Điều này giúp VeboTV trở thành một trong những sự lựa chọn đáng tin cậy nhất cho người xem. Tốc độ truyền tải trên Vebo TV thực sự ấn tượng. Chỉ cần một cú click vào liên kết, trận đấu sẽ hiển thị ngay lập tức.
Mặc dù đôi khi có hiện tượng giật lag và cần phải thay đổi liên kết, nhưng tổng thể, trải nghiệm vẫn rất tốt. Hơn nữa, sự xuất hiện của bình luận viên và khung chat trò chuyện giúp tránh được cảm giác nhàm chán khi xem một mình.
Xem bong da truc tiep trên Vebo TV là một điểm sáng trong lòng người hâm mộ. Tất cả những trận đấu từ lớn cho tới nhỏ đều được website Vebo gửi đến người hâm mộ với chất lượng Full HD cho đến 4K.
Hơn thế nữa, các thông tin, tin tức liên quan tới nhận định, cược kèo cũng được các chuyên gia biên soạn sẽ được đăng tải mỗi ngày.
Các câu hỏi thường gặp (FAQ) – Về hoạt động của Vebo
Dưới đây là những thắc mắc phổ biến mà người dùng thường gặp khi muốn hiểu rõ hơn về kênh bóng đá trực tuyến Về Bờ. Hãy tham khảo để có câu trả lời chi tiết cho mọi tình huống.
Làm thế nào để tham gia trò chuyện tại Vebo?
Trong quá trình xem bóng đá trực tuyến trên VeboTV, việc trò chuyện và thảo luận là không thể thiếu. Vebo cung cấp một khung chat ngay bên cạnh màn hình trận đấu. Người dùng có thể đăng nhập thông qua tài khoản Facebook hoặc Google để sử dụng tính năng này một cách thuận tiện.
Có phải trả tiền để sử dụng dịch vụ của Vebo không?
Như đã đề cập, VeboTV hướng đến mục tiêu trở thành kênh trực tiếp bóng đá hàng đầu miễn phí tại Việt Nam. Do đó, không có bất kỳ chi phí nào cho việc sử dụng dịch vụ và tính năng của Vebo. Tất cả các tiện ích như xem lịch thi đấu, bảng xếp hạng và tin tức bóng đá đều hoàn toàn miễn phí.

Tôi có thể đặt cược tại Vebo không?
Vebo chỉ cung cấp trận đấu trực tiếp để người hâm mộ theo dõi. Mặc dù cung cấp thông tin về tỷ lệ cược và dự đoán kết quả, nhưng Vebo không phải là nơi để đặt cược. Kênh này tập trung vào việc cung cấp một môi trường xem bóng đá lành mạnh và văn minh. Người dùng có thể tham khảo các đơn vị đặt cược mà Vebo giới thiệu.
Độ trễ giữa trận đấu trực tiếp trên Vebo và trên truyền hình là bao nhiêu?
Để tránh bị “spoiler” hoặc xem trận đấu muộn hơn, Vebo liên tục cải thiện tốc độ truyền tải để giảm thiểu độ trễ. Băng thông được mở rộng để dữ liệu trận đấu được truyền tải một cách nhanh chóng và không bị gián đoạn.
Ước tính độ trễ của Vebo so với các kênh truyền hình khác như K+, FPT là khoảng 5 – 10 giây tùy thuộc vào từng trận. Đây là con số rất thấp đối với một trang web phát sóng bóng đá trực tuyến miễn phí và có chất lượng 4K.
Lời kết
Vebo TV không chỉ là một nền tảng phát sóng bóng đá trực tuyến miễn phí mà còn là một hệ thống hoạt động với sự tận tụy và chuyên nghiệp. Mục tiêu hàng đầu của chúng tôi là mang đến sự hài lòng tối đa cho khách hàng.
Nếu bạn đang tìm kiếm một trang web để thưởng thức bóng đá mà không cần phải chi trả bất kỳ khoản phí nào, hãy truy cập ngay vào Vebo TV. Website đảm bảo toàn bộ tiện ích dịch vụ được cung cấp đều là Free (miễn phí).
